Hyperparameter optimisation for support vector machine-based disease detection in maize leaf variants
Keywords:
farmers, livestock, machine learning, RSA, SVMAbstract
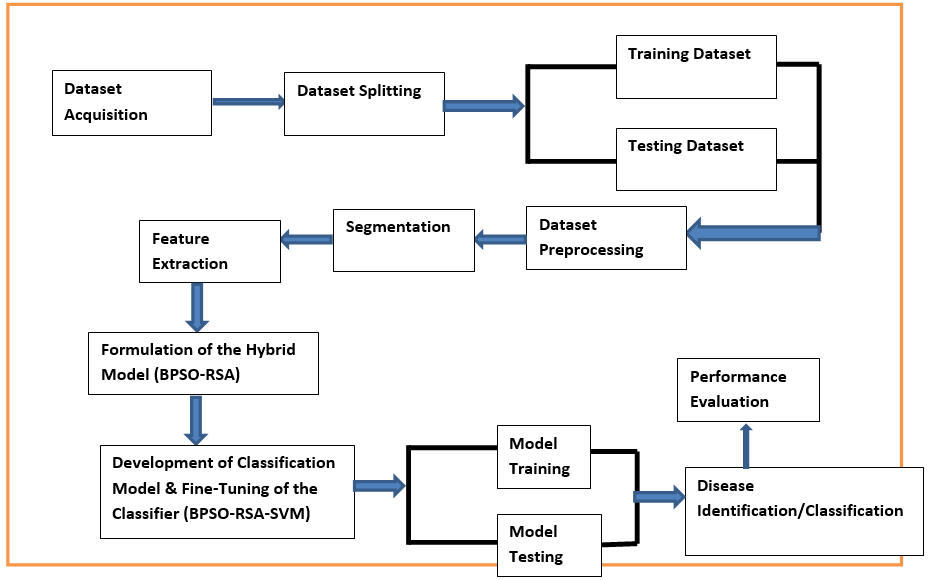
One of the main staple crops farmed and extensively consumed in Africa is maize. However, despite being widely used as a human diet and a raw material for animal feed, several diseases on leaves endanger their productivity and result in a sizable yield loss. However, Nigerian farmers typically employ antiquated techniques to detect plant diseases, which are labour-intensive and prone to mistakes, making the constant need for more effective solutions necessary. Although numerous researchers have developed classification models using Support Vector Machine (SVM) to identify and classify diseases in crop leaves. However, optimising a Support Vector Machine (SVM) is critical because it allows for fine-tuning of its parameters to achieve the best possible performance on a given dataset. Therefore, to optimise Support Vector Machines, this study created a hybrid model that combines Binary Particle Swarm Optimisation (BPSO) with a Reptile Search Algorithm (RSA). The Kaggle village datasets provided images of the leaves of maize. After being converted to grayscale, the pictures were improved with bi-histogram equalisation methods. After segmenting the leaf’s affected area using the Sobel edge detection method, texture, shape, and colour features were extracted using Gray Level Spatial Dependence and colour moment. Every classification model was trained and tested using the 10-fold approach. The performance of the suggested method was compared with a few other machine learning and deep learning models that are currently in use. Regarding identifying maize diseases, the results showed that the BPSO-RSA-SVM model performed better than all other optimised support vector machine models and some deep learning state-of-the-art models. The model demonstrates its efficiency in advancing agricultural disease detection with an average accuracy of 97.01% and a false positive rate of 3.85% compared with BPSO-SVM and RSA-SVM which achieved 96.65% & 95.44% and 3.30% & 4.60% for accuracy and false positive rate, respectively.
Published
How to Cite
Issue
Section
Copyright (c) 2025 O. B. Ayoade, M. O. Raji, A. A. Akindele, K. J. Yusuf-Mashopa, M. F. Abdulrauf, I. A. Raji, F. B. Musah

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Similar Articles
- Osowomuabe Njama-Abang, Denis U. Ashishie, Emmanuel A. Edim, Moses A. Agana, Development of a visual analogy model using transfer learning techniques , African Scientific Reports: Volume 4, Issue 3, December 2025
- Adebayo Abdulganiyu Keji, Oluwafemi Fakeye, Nneka N. Onochie, Olumide Sangotoki, Predicting long-term deposit customers using convolutional neural network and data conversion technique , African Scientific Reports: Volume 3, Issue 3, December 2024
- Gabriel James, Anietie Ekong, Aloysius Akpanobong, Enefiok Etuk, Saviour Inyang, Samuel Oyong, Ifeoma Ohaeri, Chikodili Orazulume, Peace Okafor, Enhanced machine learning model for classification of the impact of technostress in the COVID and post-COVID era , African Scientific Reports: Volume 4, Issue 1, April 2025
- Adenike Adegoke-Elijah, Theresa Omolayo Ojewumi, Kudirat Oyewumi Jimoh, ECG anomaly detection: a deep learning perspective with LSTM encoders , African Scientific Reports: Volume 4, Issue 3, December 2025
- Gabriel James, Anietie Ekong, Etimbuk Abraham, Enobong Oduobuk, Nseobong Michael, Victor Ufford, Oscar Ebong, An enhanced control solutions for efficient urban waste management using deep learning algorithms , African Scientific Reports: Volume 3, Issue 3, December 2024
- Denis U. Ashishie, Endurance O. Obi, Osowomuabe Njama-Abang, Ahena I. Bassey, Transformative approach in Lassa fever diagnostics: an innovative integrative strategy for early detection and outcome prediction , African Scientific Reports: Volume 4, Issue 3, December 2025
- A. I. Bassey, M. A. Agana, E. A. Edim, O. Njama-Abang, Intrusion detection in a controlled computer network environment using hybridized random forest and long short-term memory algorithms , African Scientific Reports: Volume 4, Issue 3, December 2025
You may also start an advanced similarity search for this article.



