GC-MS Profiling and In Silico Studies to Identify Potential SARS-CoV-2 Nonstructural Protein Inhibitors from Psidium guajava
Keywords:
Docking, GC-MS, Leaves, P. guajava, SARS-CoV-2Abstract
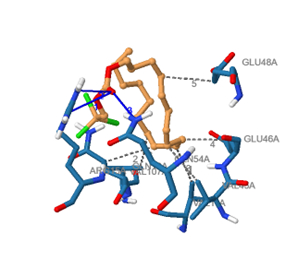
The COVID-19 pandemic, caused by the SARS-CoV-2, has prompted international concern. The aim of this study is to identify SARS-CoV-2 nonstructural protein inhibitors-potentially bioactive phytocompounds from the traditional plant Psidium guajava. GC-MS analysis of P. guajava methanol leaves was investigated. In silico molecular docking, drug-likeness, toxicity, and prediction of the compounds’ substance activity spectra (PASS) were evaluated. GC-MS analysis identified thirty (30) phytocompounds. According to molecular docking, all the phytocompounds have strong binding energies. The phytocompound beta bisabolene gave the best binding affinity of -5.0 kcal/mole. The detected compounds were all in accordance with Lipinski’s Rule of Five (RO5). This showed that the identified P. guajava compounds would have lower attrition rates during clinical trials and thus have a better chance of being marketed. According to this research, a potential COVID-19 drug could be created using the newly identified
phytocompounds of P. guajava.
Published
How to Cite
Issue
Section
Copyright (c) 2022 Ifeanyi Edozie Otuokere, Onyinye Uloma Akoh, Felix Chigozie Nwadire, Chinedum Ifeanyi Nwankwo, Joy Nwachukwu Egbucha, Chiemela Wisdom, Ogbonna Augustine Okwudiri

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Most read articles by the same author(s)
- O. U. Igwe, C. C. Oru, I. E. Otuokere, Chemical and bioprotective studies of Xylopia aethiopica seed extract and molecular docking of doconexent and cryptopinone as the prominent compounds , African Scientific Reports: Volume 3, Issue 2, August 2024



